Journal of
eISSN: 2373-633X


Research Article Volume 13 Issue 1
1UR Molecular biology of leukemia and lymphoma, Faculty of Medicine of Sousse, University of Sousse, Tunisia
2Biology department, Hail University, Saudi Arabia
3Laboratory “Valorisation of biomass and production of proteins in Eukaryotes”, Biotechnology Center, Sfax, Tunisia
4Department of Clinical Hematology, CHU Farhat Hached, Tunisia
Correspondence: Walid-Sabri Hamadou, Biochemistry Laboratory, Faculty of Medicine of Sousse, Avenue Mohamed Karoui 4000, Sousse–Tunisia, Tel (216) 73 222 600, Fax (216) 73 224 899
Received: January 30, 2022 | Published: February 28, 2022
Citation: Sawsen B, Walid-Sabri H, Nouha B, et al. Molecular characterization of Tunisian B-lineage leukemia using Kde gene rearrangements. J Cancer Prev & Curr Res. 2022;13(1):34-39. DOI: 10.15406/jcpcr.2022.13.00483
Background: Molecular approaches applied to the study of B-lineage leukemia have made significant contributions in understanding and characterizing the disease. The Kde rearrangement was targeted as clonal marker to characterize Tunisian B-chronic lymphocytic leukemia (B-CLL) and B-acute lymphoblastic leukemia (B-ALL).
Objective: We used a multiplex PCR to amplify deletional Kde rearrangements and Southern blot assay with Cκ and Kde probes in seven B-CLL and two B-ALL. Direct sequencing was performed in order to determine the n region.
Results: An interesting RSS-Kde biallelic rearrangement was found in one adult male patient with B-ALL, since only Vκ-Kde rearrangement has been reported in adult B-ALL. In B-CLL, one case with two clonal cell proliferations in favor of oligoclonality which is very infrequent in B-CLL was identified and might be associated with bad outcome. These first results need more investigation to establish correlation between molecular characteristic and the poor outcome.
Conclusion: The molecular approach was successfully improved and applied for clonality assessment of B-lineage leukemia for a better diagnosis and performing monitoring minimal residual disease.
Keywords: B-lineage leukemia, IGK gene, Kde rearrangements, multiplex PCR, chemiluminescent southern blot
The various types of B-lineage leukemia, i.e B-acute lymphoblastic leukemia (B-ALL) and B-chronic lymphocytic leukemia (B-CLL) can be considered as malignant counterparts of cells in immature and mature stage hematopoiesis respectively. The cells of the vast majority of these malignancies (98%) process the same clonal immunoglobulin gene rearrangements. Virtually all B-ALL (>95%) have rearranged IGH genes on at least one allele.1 IGK gene rearrangements and deletions occur at a relatively high frequency of 60% in precursor B-ALL,2,3 whereas the IGL rearrangements are found in ~20% of cases.4 In B-CLL the IGK gene rearrangements proceed in 30% of cases.5 As IGK gene deletion occurs when IGL gene rearrangements are produced, the deletional process is mediated via site-specific rearrangements of kappa deleting element (Kde) which is located 24 kb downstream of the Cκ constant gene segment.6-8 Two types of Kde deletional rearrangement may occur: in B-ALL, Kde rearranges in 45% of cases to Vκ gene upstream Vκ-Jκ rearrangement, thereby deleting Jκ and Cκ region, or to intron RSS located between Jκ segments ant Cκ gene, resulting in Cκ deletion in 25% of cases.3,9 The Kde rearrangements occur in approximately 50% in childhood versus 35% in adult B-ALL.5 These rearrangements may occur on one allele or both alleles in 20% and 30% of precursor B-ALL with Cκ gene or Jκ-Cκ region deletion respectively. Whereas in B-CLL Igl+, the IGK locus is 90% deleted on both alleles involving Vκ gene or RSS gene in 45% and 55% of cases respectively.5
In all cases, heptamer-nonamer recombination signal sequence is required to achieve the rearrangement. Comparable to Vκ-Jκ rearrangement, the Kde rearrangements (on Vκ gene or intron RSS) result in junctional regions in which diversity is created by trimming of the rearranging segments and insertion of template independent nucleotides by terminal deoxynucleotidyl transferase (TdT). Some diversity found in junctional sequences of Kde rearrangement has been reported in B-ALL and B-CLL. There are more insertion in N region and deletion of nucleotides found in junctional regions in B-ALL than in B-CLL.10 This might be due to continuous TdT activity during Kde rearrangement in B-ALL or less activity in B-CLL.
Kde rearrangements are specific to leukemic clone and are stable (end-stage rearrangement).11 They represent a valuable target to perform the diagnosis of B lineage leukemia (B-ALL and B-CLL) and to estimate and follow the outcome of patients by monitoring the minimal residual disease (MRD).12
In this study, the Kde rearrangement was targeted as a clonal marker to get a better characterization of nine B-lineage leukemia (7 B-CLL and 2 B-ALL). Specific Kde multiplex PCR was devoloped. We compared the results to chemiluminescent Southern blot analysis using a synthesized and cloned Kde probe and identified the junctional region for further MRD monitoring. The obtained results through this study performed for the first time in Tunisia, should influence B-leukemia’s prognostic classification in order to improve the outcome of Tunisian patients with these malignancies by assessing MRD.
Bioinformatics analysis
The bioinformatics annotation, using "the assembly GRCh 38" has been performed with three databases: NCBI, UCSC and ENSEMBL and compared to IMGT database. We have annotated in this contig: The JK segments, RS-JK, Intron RSS, the enhancer, IGKC, The intronic region, The IGK-Kde segment and RS-Kde. We have positioned the restriction enzymes: BamHI, BglII, HindIII, EcoRI, SacI, XbaI using enzymes: http://nc2.neb.com/NEBcutter2/.
Patient samples
Nine Tunisian patients diagnosed at hematology department, F. Hached Hospital of Sousse were included in this study, two patients with B-ALL (a four-year old child and a sixty-eight year old adult) as well as seven patients with B-CLL (their average age was 68 years). Informed consent was obtained from all patients according to Helsinki Declaration. The diagnosis was established according to conventional histological standards as set by the World Health Organization WHO classification and confirmed by morphological and immunophenotypic analysis. ALL samples contained 80% of blast cells. For CLL samples initial diagnosis was based on blood lymphocyte count >4×109/l with mature appearing lymphocytes and 40% of lymphocyte in bone marrow aspirates.
Peripheral blood samples were obtained from patients at diagnosis with high blastosis (>75%) to investigate detectable IGK-Kde rearrangements. A control population was recruited among healthy blood donors. Blood samples were obtained after the donors had given their informed consent.
Mononuclear cells and DNA isolation
Mononuclear cells (MNC) were isolated by Ficoll-Paque density centrifugation and were directly used for DNA isolation. DNA was extracted with phenol chloroform. DNA was quantified by spectrophotometry assay and tested for quality on 0.7% agarose gel electrophoresis. The control DNA was obtained from MNC of healthy volunteers.
Southern blot analysis
Southern Blot was performed to investigate the presence of IGK-Kde rearrangement (Figure 1a). DNA (10 µg) was digested with appropriate restriction enzyme (BamHI, BgIII, HindIII) (Boehringer Mannheim), size separated on a 0.8% agarose gel and transferred on to nylon membrane (Hybond N+, Amersham Pharmacia, Biotech). The hybridization signal was detected by CDP-Star chemiluminescent system (Amersham Pharmacia, Biotech) as previously described.13 The IGK-Kde gene configuration was analyzed by mean of Alkalin phosphatase labeled IGK-Cκ or IGK-Kde probes.
IGK probes
The IGK-Cκ probe, clone R17CK, is a 2.5 Kb EcoRI fragment containing the IGK constant region cloned in pUC.14 The IGK-Kde probe is specific to 5’ region of Kde segment which is located downstream of the Cκ gene. It was amplified by PCR in the mixture containing: 200 ng of genomic DNA of healthy volunteer, 1x PCR buffer, 1.3 pmol of upstream and downstream specific primers of the Kde segment, 200µM of dNTP mixture, 1.5mM MgCl2 and 2.5 U of Go Taq polymerase (Promega) in a final volume of 50µl. The amplified Kde probe was purified by Qiagen kit purification (QIAquick PCR Purification Kit) and cloned using the vector pCR®2.1 (TA cloning kit by Invitrogen) in Top10 rubidium competent cells. After blue-white screening on X-Gal/IPTG indicator plates, plasmid DNA was isolated using alkaline lysis method. The plasmid DNA recombination was confirmed by appropriate EcoRI restriction enzyme digestion and PCR amplification.
PCR analysis
Clonal IGK-Kde gene rearrangements were investigated by Simple and Multiplex PCR amplification. A reverse Kde primer was used in combination with one of six VK family-specific primers or with RSS primer located at the JK-CK intron. These PCR were performed according to BIOMED 2 consortium.5,15 All PCR analysis were realized in triplicate with a positive control (DNA used as standard in which the deletional rearrangement is already defined). The amplification products were electrophoresed 2% agarose gel and stained with sight DNA stain (Bioatlas). All obtained rearrangement is confirmed by simple PCR.
Sequence analysis
Junctional region sequence was obtained by direct cycle sequencing using BigDye® Terminator v3.1 Cycle Sequencing Kit and an ABI Prism 3730xl DNA Analyzer automated sequencer (Applied Biosystems, USA). Sequence reactions were performed using sequence primers of VκI, RSS and Kde according to the manufacturer’s instructions. The junctional region of Kde rearrangement was sequenced twice, starting from two independent PCR reactions and from opposite sequence orientations. The sequence was analyzed using V-quest IMGT (http://www.imgt.org/IMGT_vquest/vquest) and confirmed with Ig-BLAST sequence similarity searching tool http://www.ncbi.nlm.nih.gov/igblast/
Bioinformatics analysis
The bioinformatics analysis permitted to localize the five IGKJ families and their respective restriction sites, also for RSS introns, the enhancer, the constant region IGKC and Kde segment. All restriction enzymes sites were subsequently positioned and visualized.
PCR analysis
We synthesized Kde probe for optimal detection of IGK rearrangements by Southern blot assay (Figure 1a). We amplified by PCR one fragment with the expected size as it was controlled by 2% agarose electrophoresis of the purified fragments. The sequence of the Kde fragment was verified by direct sequence and blasted in NCBI (locus X03957) (data not shown). The 536 pb fragment is the IGK-Kde probe which is specific to 5’region of Kde segment located downstream of the Cκ. This probe was cloned in the vector pCR®2.1 and confirmed by enzymatic digestion, PCR amplification and direct sequence.
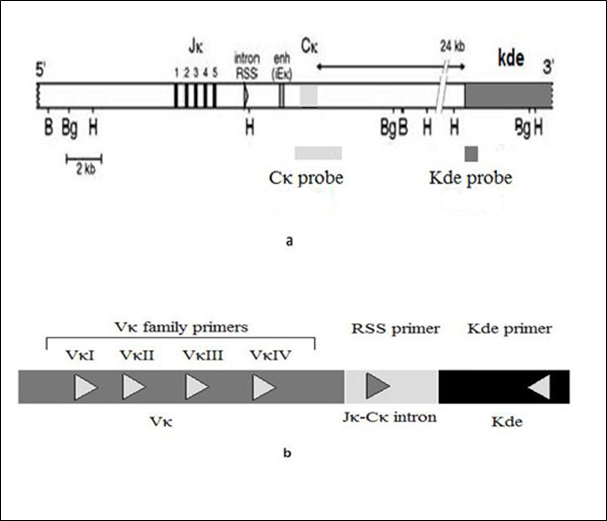
Figure 1 (a) Schematic location of Cκ probe (2.5 kb) and Kde probe (536 pb) for Southern blot analysis. (b) Primers sequences and location for Kde clonal rearrangements PCR analysis.
Southern blot
The IGK locus was investigated by Southern blot analysis in 2 B-ALL and 7 B-CLL at diagnosis for the presence of detectable IGK-Kde rearrangement. The IGK locus was analyzed with BamHI and HindIII digest hybridized to a Cκ probe, HindIII or BglII digest hybridized to a new synthesized Kde probe.
B-All
In one adult pre B-ALL (sample 1), the southern blot analysis showed neither germline nor rearranged band with BamHI digest and Cκ probe, the IGKC gene was then deleted on both alleles (Figure 2a). Furthermore, the new Kde probe detected only one 4.3 kb BglII and one 7.8 kb HindIII rearranged bands (Figure 2b, c). No germline band was present in both digests. These Kde rearrangements occur on both chromosomes even though one rearranged band was revealed probably due to co-migration of the two BglII or HindIII rearranged fragments. This adult B-ALL had biallelic IGK-Kde rearrangement.
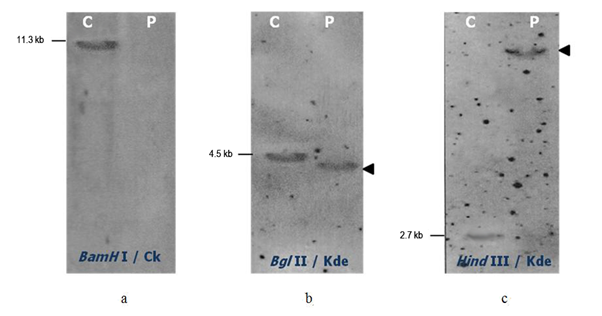
Figure 2 Southern Blot analysis of the IGK gene in adult pre-B-ALL (sample 1). Hybridization of alkaline phosphatase Cκ or Kde probe to the BamHI (a), BglII (b) or HindIII (c) digested control DNA (C) and DNA sample from pre B-ALL patient (P). The size of germline bands are given on the left and the assignment of the rearranged bands by an arrow on the right.
In children B-ALL (sample 2) we didn’t find any Kde rearrangement by Southern blot. The IGKC was in germline configuration.
Using five PCR primers (VκI to VκIV and intron RSS) in five single PCR, we amplified in the adult pre B-ALL (sample 1) a discrete band at the expected 480 pb size corresponding to the RSS rearrangement. The same result was obtained with the five PCR primers combination in the multiplex PCR essay (Figure 3a). This pre B-ALL shows the intron RSS-Kde rearrangement as deletional rearrangement. In the children B-ALL, no amplification was obtained confirming the Southern blot results.
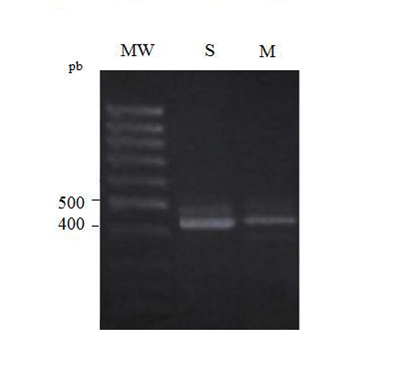
Figure 3 (a) 2% Agarose electrophoresis of simple (S) and multiplex (M) PCR IGK-Kde products in pre B-ALL case (sample 1); MW: molecular weight. (b) 2% Agarose electrophoresis of simple (S) and multiplex (M) PCR IGK-Kde products in B-CLL cases (samples 3, 4, 5, 7) - MW: molecular weight.
B-CLL
In B-CLL cases the Kde was rearranged on one allele (sample 3) or both alleles (samples 4, 5, 6, 7) as it was deduced by BamHI digest with Cκ probe and confirmed in BamHI, BglII and HindIII digests with Kde probe (Figure 4). In multiplex PCR assay we have amplified the Kde rearrangement that involves VκI, VκII and RSS genes (Figure 3b). In B-CLL (sample 6) the chemiluminescent Kde probe revealed two rearranged bands in the Southern blot analysis with the persistence of the germline one (Figure 4c). In this case multiplex PCR amplified two deletionel Kde rearrangements involving VκI and RSS genes. In two B-CLL (samples 8, 9), the IGK-Kde locus was in germline configuration as observed by Southern blot analysis with Kde probe. This result was confirmed by BamHI digest using Cκ probe, as two new bands were obtained in favor of IGKC gene rearrangement on both alleles (Figure 4a, c). The multiplex PCR assay did not reveal any rearrangements.
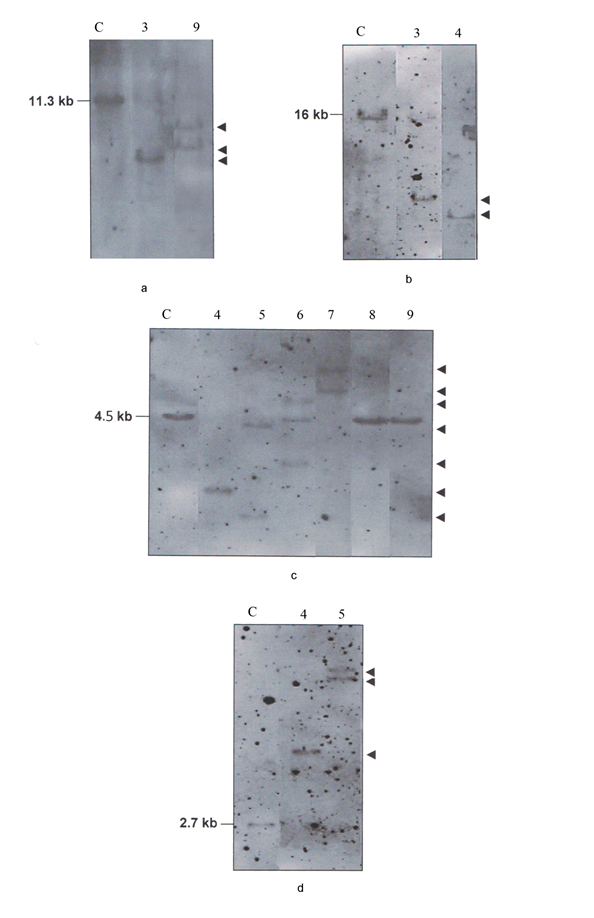
Figure 4 Southern Blot analysis of the IGK gene in B-CLL cases (samples 3, 4, 5, 6, 7, 8, 9). Hybridization of alkaline phosphatase Cκ probe to the BamHI digest (a). Hybridization of alkaline phosphatase Kde probe to the BamHI digest (b), BglII digest (c), HindIII digest (d). C: control DNA. The size of germline bands are given on the left and the assignment of the rearranged bands by an arrow on the right.
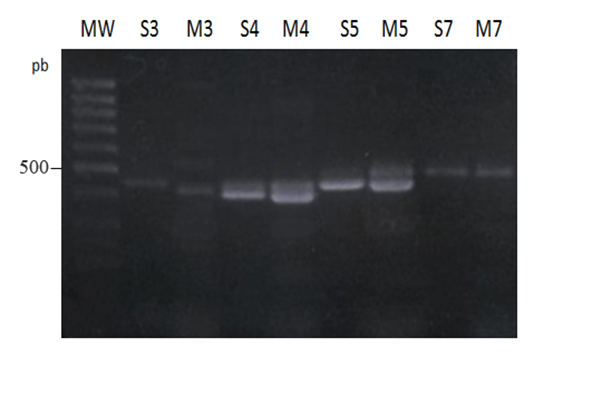
Figure 5 2% Agarose electrophoresis of simple (S) and multiplex (M) PCR IGK-Kde products in B-CLL cases (samples 3, 4, 5, 7) - MW: molecular weight.
Sequence analysis
The Kde rearrangements were sequenced in pre B-ALL (sample 1) and B-CLL (sample 5, sample 6). The result of the N-region sequenced confirmed the occurrence of the Kde segment deletion. In pre B-ALL with RSS-Kde rearrangement, the number of inserted nucleotides in the junctional region was (-3/+4/0). For B-CLL (sample 5) with bi-allelic Kde rearrangement, the first one was VκI-Kde with N-region of (-6/+2/0), and the second was RSS-Kde characterized by (0/+1/0). For B-CLL (sample 6) with VKI-Kde rearrangement the number of inserted nucleotides in the junctional region was (-3/+1/-2).
Since approximately 50% of precursor B-ALL and 30% of B-CLL display mono or biallelic IGK-Kde rearrangements.3,15 We investigated the Kde deletion as an independant diagnostic and prognosis tool for Tunisian B lineage leukemia. We have developed in our laboratory a chemiluminescent Southern blot assay to determine lymphoproliferation clonality using a set of immunoglobulin Ig probes including the Ig light chain Cκ one.13 However, Cκ probe only deduces the deletional rearrangement of IGK gene whereas the more specific Kde probe allows the detection and confirmation of IGK deletion. Therefore, we have developed, cloned and used a specific Kde probe to precise clonality and diagnosis of B-ALL and B-CLL.
For B-ALL cases analyzed in this study (Table 1), the adult pre B-ALL is characterized by the expression of the Igl light chain which is in favor of the Igκ light chain deletion. Indeed, this case displays a biallelic IGKde rearrangement as seen with the new cloned Kde probe in BglII and HindIII digests that confirms biallelic deletion observed also with Cκ probe in Southern blot analysis. The multiplex PCR assay precises the RSS-Kde rearrangement on two chromosomes. In the children B-ALL, the IGK locus is in germline configuration as deduced by Southern blot and PCR analysis. The investigation of the IGH locus by Southern blot in three BamHI, EcoRI and HindIII digests, revealed the presence of biclonal malignant proliferations (data not shown). Oligoclonality has been reported in 30-40% of B-ALL and may be a bad prognosis factor.16
|
Patient |
Sex |
Age |
Diagnosis |
Southern blot |
PCR Multiplexe |
|
|
IGK configuration (Cκ probe) |
IGKde configuration (Kde probe) |
|||||
|
1 |
M |
68 |
Pre B-ALL |
D/D |
R/R |
RSS-Kde |
|
RSS-Kde |
||||||
|
2 |
M |
4 |
B-ALL |
G/G |
G/G |
- |
|
3 |
F |
63 |
B-CLL |
R/D |
R/G |
VκI-Kde |
|
4 |
M |
62 |
B-CLL |
D/D |
R/R |
VκII-Kde |
|
VκII-Kde |
||||||
|
5 |
M |
82 |
B-CLL |
D/D |
R/R’ |
VκI-Kde |
|
RSS-Kde |
||||||
|
6 |
M |
61 |
B-CLL |
D/G |
R/R’/G |
VκI-Kde |
|
RSS-Kde |
||||||
|
7 |
M |
66 |
B-CLL |
D/D |
R/R’ |
RSS-Kde |
|
8 |
M |
70 |
B-CLL |
R/R’ |
G/G |
- |
|
9 |
F |
70 |
B-CLL |
R/R’ |
G/G |
- |
Table 1 Southern blot and PCR results in B-ALL and B-CLL cases
M, male; F, female; G, germline band; R R, rearranged band; D, deleted locus; -, absence of IGKde rearrangement
In this study, the result of the Southern blot revealed with the new Kde probe a new germline restriction fragment of 4.5 kb in BglII digest instead of the 7.6 kb expected size described in the literature.3 This might be due to a polymorphic BglII restriction site upstream on the Kde segment. This restriction fragment length polymorphism will be further investigated.
For seven B-CLL cases (Table 1), only two displayed the Kde locus in germline configuration as they have rearranged the IGK on both chromosomes like 20% of B-CLL Igκ+. Southern blot and multiplex PCR results revealed similar results. All other five B-CLL displayed mono or biallelic Kde rearrangements as concluded by common results to multiplex PCR and Southern blot using Cκ or Kde probes. Multiplex PCR has deduced on VκI, VκII and RSS genes involved in Kde rearrangement. In four B-CLL, VκI, VκII genes were mostly used as described.5 In one B-CLL (sample 6) the Southern blot results has concluded to two clonal cell proliferations. The two amplified VκI-Kde and RSS-Kde rearrangements belong probably to two independent clones. The oligoclonality is very infrequent in B-CLL and might be associated with bad outcome. This result highlights the complementarities of the Southern blot and PCR assays in Kde rearrangement analysis especially when more than one clonal proliferation is present in the leukemia.
Multiplex PCR confirms not only the Kde locus deletion but also the clonality of the hematological malignancies determined already by the Southern blot analysis. However, PCR is not suitable for detecting biclonal Kde rearrangement. We confirm that the Southern blot is more accurate and necessary to get a better characterization of B lineage leukemia.3 The two methods are complementary. In B-CLL (sample 7), the Southern blot and PCR were not in agreement as we detected biallelic Kde rearrangement by Southern blot but rather only monoallelic RSS-Kde rearrangement by multiplex PCR. This may be due to the involvement of other Vκ gene family not included in this study like (VκVII) or non-specific Kde rearrangement previously described like Jκ-Kde rearrangement.17
PCR based techniques offer a good alternative for clonality assessment. Particularly, the multiplex PCR represents a rapid and highly specific method for detection and identification of the most common deletional rearrangement of Kde.15 The primers conception allows identifying the majority of the rearrangement in one reaction and PCR products are easily assigned to a specific rearrangement by a size ranged from 326 pb to 483 pb. Therefore, we have used in this study the Kde multiplex PCR assay as it provides a reliable and easy tool for establishing clonality in lymphoid malignancies and confirming diagnosis in conjunction to the morphological and immunophenotypic data. This method, in addition with Southern blot analysis, might be helpful when standard techniques are equivocal and fail to determine the diagnosis. Hence, we have improved the molecular approach we developed in Tunisia and successfully applied it to clonality assessment of B lineage leukemia.
We have determined by single and multiplex PCR in pre B-ALL the occurrence of intron RSS-Kde rearrangement. Previous study has reported that Kde deletion in adult B lineage ALL involves only Vκ-Kde rearrangement and did not detect any RSS-Kde rearrangement which shows a difference between children and adults B-ALL.5,18 In our study, we identified a biallelic RSS-Kde rearrangement in one adult male patient with B-ALL and confirmed the result of Nakao.19 He had detected six RSS-Kde rearrangements in five adult B-ALL patients and he noted that the frequency of this rearrangement in adult B-ALL is compatible with the incidence in childhood B-ALL. We are investigating more adults B-ALL cases for further characterization of RSS-Kde rearrangement and possible correlation with prognosis since the pre B-ALL patient studied relapsed.
Sequence analysis of Kde rearrangement allows describing the N-region. For pre B-ALL the number of inserted nucleotides in the junctional region was (0/+6/0) whereas in B-CLL (sample 5) in the VκI-Kde rearrangement the N-region is (-6/+2/0), and the second RSS-Kde rearrangement the N region is (0/+1/0). We have confirmed the relatively large N-region found in precursor B-ALL comparatively to chronic B cell leukemia10 due to continual activity of TdT during the rearrangement of Kde segment.20 As Kde rearrangements are stable during the disease course, the junctional N-region is specific and stable target for minimal residual disease monitoring which we are performing for complete molecular characterization and improvement of outcome in B leukemia Tunisian patients. In other study, we performed real time quantitative PCR using allele-specific oligonucleotide targeting N-region in B-ALL cases, and prominent results were obtained for MRD monitoring.21 Further investigation on B-CLL are needed. Nowadays serval markers have been performed for MRD assessing, such as IKZF1 deletions or NOTCH1/FBXW7 mutations but Kde rearrangement remain a valuable tools in case of absence of mutations or deletions.
These findings could be a valuable for better follow up of patients by assessing minimal residual monitoring targeting light chain rearrangement for patients with high risk of relapse as well identification of prognosis risk group and orientation for adequate therapeutic decision.
We are grateful to Professor Marie-Paule Lefranc from IGH of Montpellier (France) for her kindly gift of the Cκ probe. This work was supported by the Ministère de l’Enseignement Supérieur, de la Recherche Scientifique et des Technologies de l’Information et de la Communication of Tunisia.
No conflicts of interest between any authors have been noticed.

©2022 Sawsen, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.